Bacterial pathogens use host as substrates for their proliferation. They thus selected, along evolution, molecular mechanisms allowing them to colonize, feed and proliferate on or inside host organisms. Some bacterial pathogens (like Salmonella or Shigella) are very well characterized, but some bacteria causing worldwide diseases are still poorly understood. It is the case of Brucella, a genus responsible for brucellosis, a major zoonosis affecting many mammals, including domestic animals like sheep, goats and cows. Two research groups are active on Brucella in our research unit.
Brucella belong to the alpha-proteobacteria group, a taxon comprising the bacterial model of differentiation Caulobacter crescentus. C. crescentus is known to be asymmetrically organized at the cellular and molecular levels. In our group, we mainly concentrate our studies on Brucella abortus, a species essentially found in cows infections.
Other the last years, we made the following discoveries :
• B. abortus shares many cell cycle regulators with C. crescentus and other alpha-proteobacteria
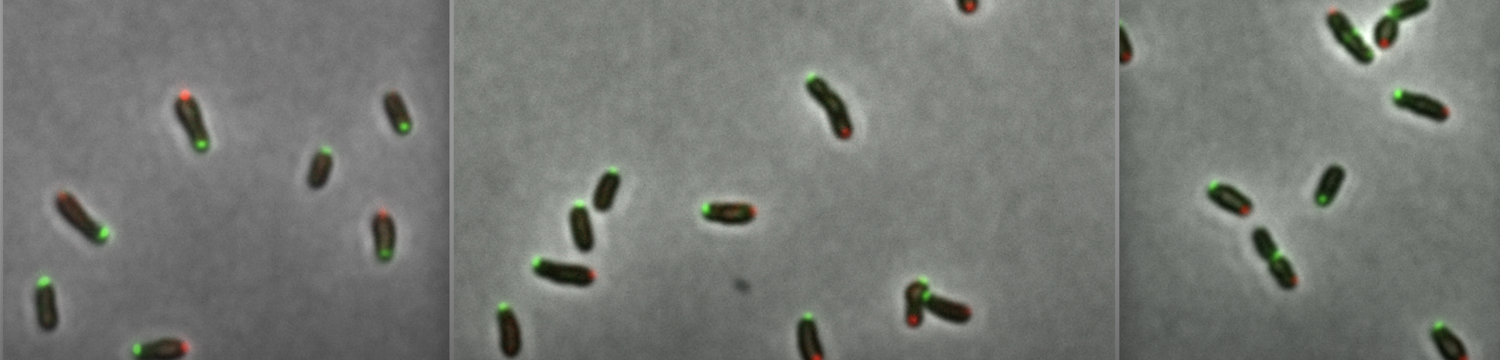
• B. abortus cells are asymmetric, the two poles (new pole and old pole) can recruit different proteins
• When B. abortus enters a host cell in a simplified model of infection, one stage of the bacterial cell cycle (G1) is favored for internalization, and bacteria can remain several hours in a non-growing, non-replicating stage
• During their intracellular traffic, B. abortus suffers from starvation and alkylating stress
• B. abortus outer membrane contains beta-barrel proteins that are covalently attached to the peptidoglycan through their N-terminal extension
• B. abortus envelope (including lipopolysaccharide, peptidoglycan, and at least two outer membrane proteins) grows through the new pole and at the division site
• Lipopolysaccharide is decorated with an O-chain (or O-antigen) by a bifunctional O-antigen ligase in the periplasm, and exported to the outer membrane and the new pole or the division site thanks to localized Lpt proteins (LptB2CFG) at these specific sites
Our current research is an integrated approach, studying basic molecular biology of the Brucella abortus pathogen, including :
- envelope structure, biosynthesis, and its regulation during polar growth
- aggressions, feeding, and starvation in culture and inside host cells
We use genetic approaches such as deletion strain generation and characterization, genome-wide transposon mutagenesis followed by deep sequencing (Tn-seq), and generation of genetically engineered strains to follow protein localization in B. abortus.